Table Of Content
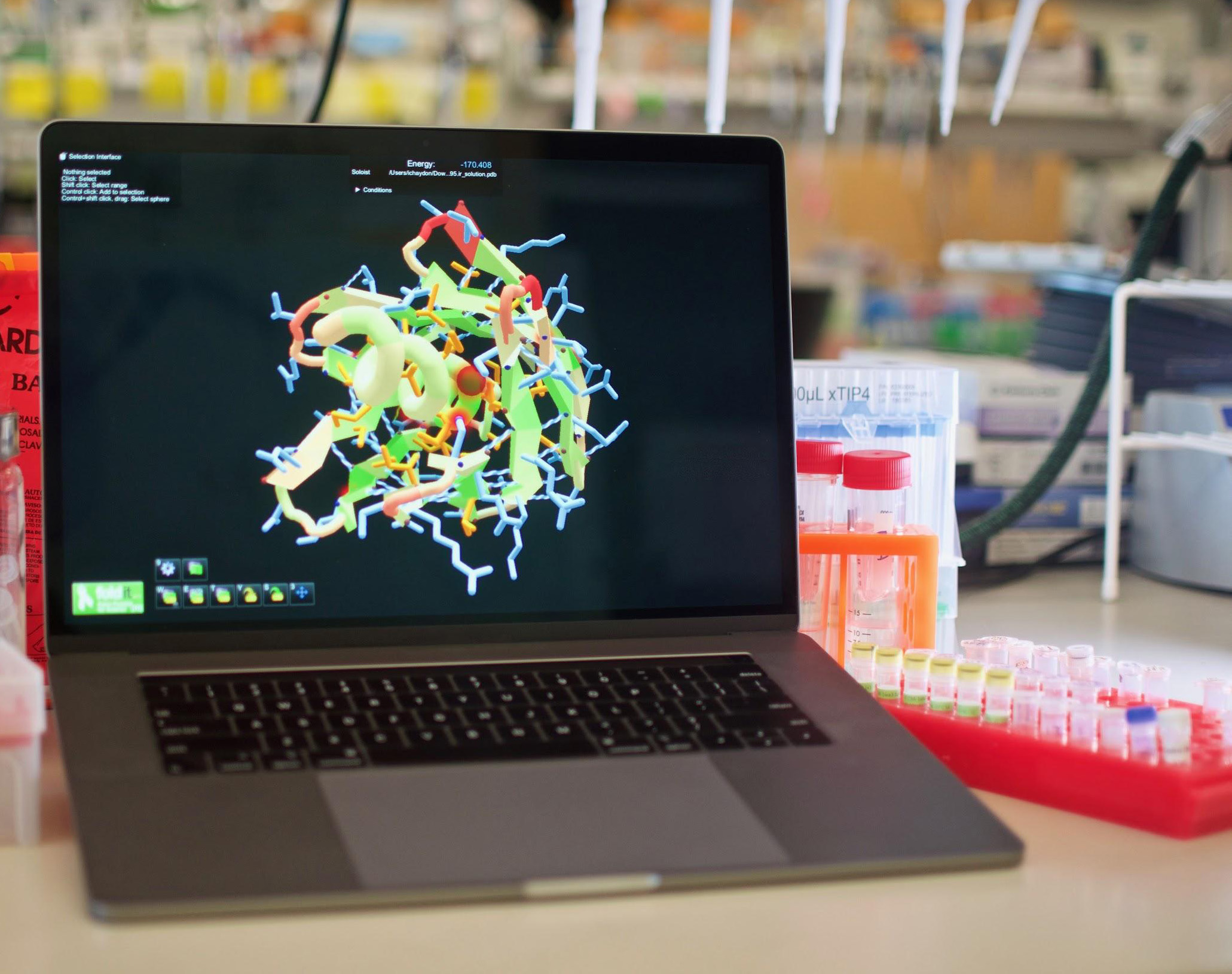
On another level, the mission of the lab is training the next generation of scientists and leaders in the field of regenerative medicine. The lab is committed to generating the best work environment to make the best science possible, and to foster multidisciplinary research using expertise from engineering, developmental biology, medical science and computer science. Besides, they catalyze rapid chemical reactions in mild conditions such as standard pressure and non-toxic solvents. With such attractive properties, it is no wonder Biochemists and Chemists have dreamed of designing proteins with customizable properties for decades.
Students and Postdocs
A great label shows the world what you stand for, makes people remember your brand, and helps potential customers understand if your product is right for them. Labels communicate all of that through color, typography and other design elements. A bold label design for a plant protein would likely involve bright, eye-catching colors and typography to capture consumers' attention. The design would also need to clearly communicate the benefits of the plant protein and appeal to consumers looking for healthy, sustainable protein options. The result would be a striking label design that stands out on the shelf and effectively communicates the product's value to consumers.
Baker Lab
The Course: Biology Design to Data - UNLV NewsCenter
The Course: Biology Design to Data.
Posted: Thu, 26 Oct 2023 07:00:00 GMT [source]
By leveraging the power of computational modeling, our researchers are creating solutions to some of the most pressing challenges in medicine, technology, and sustainability. Advanced computational techniques, including novel machine learning algorithms, allow our scientists to model the behavior of proteins at the atomic level. This knowledge helps researchers generate novel proteins with optimized stability, binding affinity, or catalytic activity. The principles of physics play a crucial role in protein design, particularly in the development of computational models and simulations. By applying concepts from statistical mechanics, thermodynamics, and molecular dynamics, researchers can predict how a protein will fold and interact with other molecules in its environment.
Chun-Chen Jerry YaoGraduate Student
For larger complexes involving multiple different subunits, the team was able to detect the configuration of the proteins in the complex using a mass spec technique called surface-induced dissociation. By slamming the molecules into a surface to break apart the subunits and then measuring which subunits colocalized among the resulting fragments, they could see how the subunits were arranged in the large complex. This type of structural characterization provides critical information to verify whether synthetic protein complexes are forming the way the designers expect. Today IPD lists seven ongoing projects to address the pandemic, from designing nanoparticle vaccines and anti-inflammatory proteins to screening existing drugs. Through Baker’s hypercollaborative nature and his desire to distribute the technology for broad adoption, de novo protein design may one day become a part of every protein engineer’s toolbox. Ben graduated from Johns Hopkins University with his Bachelors in Biophysics and a minor in Computer Science.
Machine learning for functional protein design
PvP was the first company launched through the University of Washington’s Translational Investigator Program. Takeda Pharmaceuticals fronted the money to conduct phase 1 clinical trials on the novel enzyme, called KumaMax, and in February 2020 exercised its option to acquire PvP. The University of Washington’s Institute for Protein Design has become a hub to galvanize de novo protein engineering, citizen science and much more since its founding in 2012. Bioactive PeptidesChemically synthesized molecules with predictable structures and functions. Advanced Drug DeliveryNanoscale protein assemblies that move therapeutics to specific cells within the body.
AI designs new drugs based on protein structures
Mother Nature has created some incredible tools, but those solutions have come about as a result of evolutionary pressures, including the need to conserve genetic real estate by making proteins that serve multiple functions. Whereas a natural protein may be the solution that evolution has arrived at, it may well not be the most efficient tool for a task thought up by a human. Creating new proteins, Baker says, allows the tools to be specifically directed, as well as modular and customizable to other uses. El-Samad’s return to San Francisco commenced “an intense and beautiful” collaboration, in which Baker’s team would send her computationally derived protein sequences and her lab would synthesize the proteins and test them. “It was very clear from the get-go that thing was going to work,” recalls El-Samad.
Future Food-Tech Alternative Proteins
During undergrad, she worked on molecular immunology at JHU and machine learning for protein engineering at Microsoft Research. Jody is working on de novo design of ligand-binding proteins using computational and deep learning approaches. Our lab invents and uses synthetic biology tools and approaches to build tissues in the lab. Over the past 16 years, UW researchers have made significant progress in protein design and protein structure prediction, developing the world leading Rosetta software. Over this period, UW scientists have developed methods for designing proteins with a wide range of new functions, including catalysts for chemical reactions, HIV and RSV vaccine candidates, and flu virus inhibitors. The IPD integrates these strengths in protein design with Seattle-area expertise in biochemistry, engineering, computer science and medicine, and leverages the exceptional Seattle strength in the software industry.
Jio JeongRotating Graduate Student
In February, Rosetta successfully predicted the 3D structure of the SARS-CoV-2 spike protein, which allows the virus to gain access to human cells. The flurry of effort that accompanied the project crashed some of the networked computers, but with the protein structure now in hand, work is underway to fight the disease. Researchers crafted mini-protein binders that perfectly complement the spike protein, preventing it from latching onto cellular receptors. To make a vaccine, IPD researchers are attaching the spike protein to a synthetic virus-like particle. The particle displays the spike protein in a repetitive array, an arrangement designed to stimulate a vigorous immune response. Researchers at the University of Washington’s Institute for Protein Design (IPD) don’t like to work alone.

Jerry is a graduate student in Harvard’s Molecules, Cells, and Organisms (MCO) program. During his gap year before starting at Harvard, he served the Taiwanese army and then worked with Dan Tawfik at the Weizmann Institute to investigate the origin of peptides and the genetic code in the origin of life. For his PhD, he hopes to elucidate the principles of biomolecular evolution using protein design, evolutionary biology, and high-throughput experimental approaches. When he is not thinking about science, he enjoys cooking, hiking, backpacking, learning languages, and exploring the evolution of human artifacts. Kevin is an undergraduate at Harvard studying computer science and molecular & cellular biology.
The Cochran Laboratory uses interdisciplinary approaches in chemistry, engineering, and biophysics to study and manipulate complex biological systems, with a focus on developing new technologies for basic science and biomedical applications. In addition, combinatorial and rational methods are used to engineer designer protein and peptide ligands for a variety of applications including wound healing, cardiac tissue engineering, and cancer imaging and therapy. Until recently, however, the discovery of binding proteins depended on the serendipity of high-throughput experimental screening and animal inoculations.
"Our work has made the world of proteins accessible for generative AI in drug research," Schneider says. "The new algorithm has enormous potential." This is especially true for all medically relevant proteins in the human body that don't interact with any known chemical compounds. Protein design weaves together principles from biology, chemistry, and physics, allowing researchers to create novel molecules with remarkable precision and functionality. In recent years, machine learning has proven to be a powerful way to model and design proteins on the computer. Rather than having human experts try to encode their knowledge into software, this strategy allows computers to train themselves to detect patterns in proteins. Recognizing the impact of this innovation, the journal Science dubbed the application of machine learning to protein science their 2021 Breakthrough of the Year.
No comments:
Post a Comment